Clinical Trials Are Drowning in Data but Starving for Patients
Building a public search tool for 500,000 clinical trials
The first clinical trial happened on a damp naval ship in 1747. James Lind, a Scottish naval surgeon, divided twelve scurvy-stricken sailors into pairs to test different treatments: oranges and lemons, cider, vinegar, sulfuric acid, garlic paste, and seawater.
At the time, scurvy was devastating naval forces. Naval historian Nicholas Rodger wrote "It has been seriously suggested that a million British seamen died of it in the eighteenth century – a figure which implies that everybody who served in the Navy died of scurvy approximately twice." Even though Lind ran out of citrus after six days, the sailors eating lemons recovered quickly enough to care for their companions still suffering through sulfuric acid treatments.
Today, we run 74,000 clinical trials simultaneously. Finding the right patients is considerably harder than rounding up sailors with scurvy – and we're a bit pickier about sample sizes.
The Trial Matching Landscape
The business of finding patients for clinical trials is massive. Pharmaceutical companies spend billions annually on clinical trials, with a significant portion going to Contract Research Organizations (CROs) and trial operators who handle patient recruitment. These operators can charge tens of thousands of dollars per enrolled patient, making successful trial matching incredibly valuable.
But recruitment challenges are widespread: approximately 80% of clinical trials are delayed or closed due to recruitment problems, with delays costing sponsors millions. Even when trials do launch, 37% of sites under-enroll and 11% fail to enroll a single patient.
For patients, particularly those with cancer, clinical trials can offer access to potentially more effective treatments. Since the trial sponsor covers costs, trials provide a route to cutting-edge therapeutics that might otherwise be inaccessible, especially where public health systems don't cover newer treatments.
Trial matching grows more complex as medicine advances:
Precision Medicine: As treatments become more targeted to specific genomics, eligibility criteria become increasingly specific. This shrinks the potential participant pool dramatically, forcing operators to search harder for qualified patients.
Geographic Complexity: Modern trials recruit internationally, with decentralized setups where no single site hosts all participants. Operators coordinate across multiple locations while maintaining consistent protocols.
Diagnostic Barriers: Many eligibility criteria require biomarker or genetic tests that aren't part of standard workups. Trial operators often need to fund additional testing just to determine if someone qualifies.
The value and complexity of this problem should make it attractive to startups. But despite the massive opportunity, it's become a graveyard for new companies due to complex enterprise sales cycles, siloed patient data, and complicated stakeholder relationships (read: healthcare). LLMs have started tackling pieces of the problem (like parsing eligibility criteria), but we're still far from a complete solution.
A Digital Registry
ClinicalTrials.gov was created in 2000 as a repository for clinical trials, both in the U.S. and internationally. Later in 2007, it became mandatory for any clinical trial operating in the U.S. to register on this site, dramatically expanding its scope. Each listing typically includes:
• Title and study description
• Trial type and phase
• Locations (sites) administering the trial
• Eligibility criteria, specifying who can and can’t participate
The registry now contains over 500,000 trials and sees millions of visitors each month — it's become the global standard for trial registration and transparency. Researchers, clinicians, and the public can freely access detailed information about almost any intervention being tested.
There's no shortage of AI trial matching software, but almost every solution I found lives behind enterprise walls, only accessible through specific health systems. This makes business sense - these tools need to integrate with medical records and often require complex infrastructure. But it also means we can't compare how well different approaches work, and leaves patients and doctors outside those systems without access to these tools. It's frustrating — there's been massive effort to make trial data public through ClinicalTrials.gov, but the tools to make that data truly accessible aren't available to most people who need them.
I wanted to explore how modern AI search and reasoning techniques could help make this public data more accessible. The first challenge was improving on ClinicalTrials.gov's search functionality.
Semantic Search
ClinicalTrials.gov has basic keyword search, but that's not enough - when you search for "metastatic melanoma," you should find relevant trials even if they use different technical terms like "stage IV melanoma" or describe the condition in other ways. Embeddings make this semantic search possible by mapping text into a space where similar meanings sit close together, regardless of the exact words used.
Mapping Searches to Trial Structures
When we embed text into a high-dimensional vector space, semantic similarity translates into geometric proximity. But there's a nuance: even a clinically detailed query like "seeking Phase 2 immunotherapy trials for metastatic melanoma" doesn't naturally map to how trials are structured in the database. The search “looks” different from the documents being searched.
Clinical trials follow strict documentation patterns. A single concept might appear differently across a trial's listing:
The title follows a rigid academic format ("A Phase 2 Study of Anti-PD1 Therapy in BRAF-mutated Melanoma")
The summary uses standardized medical terminology and trial conventions
The detailed description contains background, methodology, and rationale
To bridge this gap, I use hypothetical documents - imagined versions of how the user's query could appear if it were actually a clinical trial. When someone searches, the system generates multiple variations that mirror real trial documentation: a title version, a summary version, a detailed description version, and an eligibility criteria version.
By embedding these hypothetical documents alongside our actual trial database, we can project the user's query into the same spaces where trial information naturally lives.
The final search uses a weighted combination of these embeddings. This helps ensure we catch relevant trials regardless of where and how they encode the matching criteria - whether it's in the technical language of the title, the structured format of the eligibility criteria, or the detailed methodology sections.
Visualizing Trial Similarity
The embeddings we generate live in hundreds of dimensions - far more than we can visualize directly. To help validate how our search system is working, I use UMAP to project these high-dimensional vectors down to 3D space. While this reduction loses some nuance, it gives us a way to sanity check how different trials cluster together.
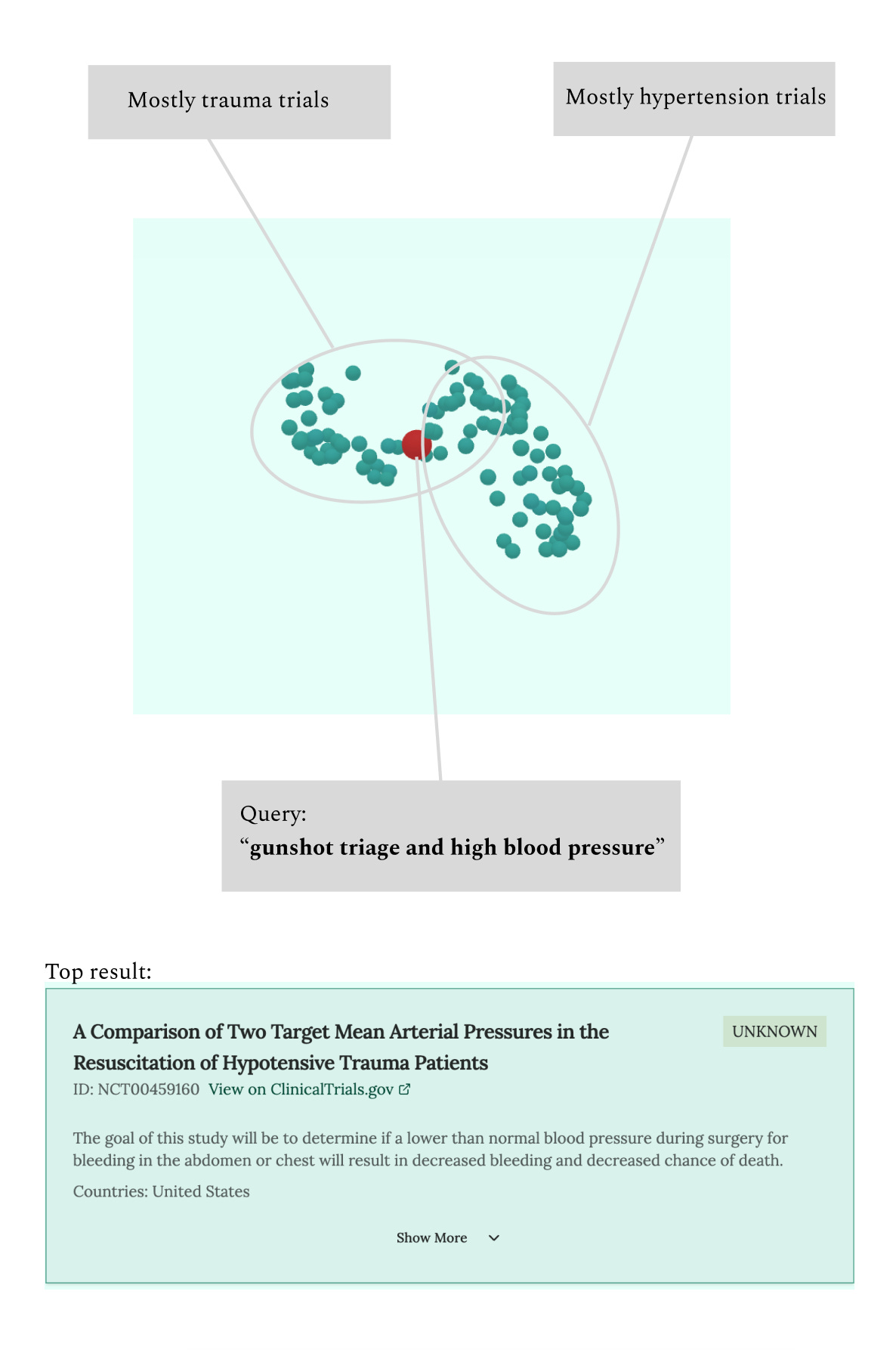
You can explore these visualizations in the tool by toggling "Show 3D trial view". The visualization shows your search query (projected through the hypothetical documents) as a point in space, surrounded by the nearest clinical trials. Different types of queries create distinctly different patterns:
A focused query like "breast cancer" creates a tight cluster - you'll see trials grouped closely together, with related terms like "breast carcinoma" appearing nearby. This clustering suggests our hypothetical documents are successfully replicating the standardized language used in oncology trials.
More complex queries reveal interesting patterns. Search for "gunshot triage and high blood pressure" and you'll see the query sitting between two distinct clusters: trauma-oriented trials on one side and cardiovascular trials on the other. The most relevant trials appear at the intersection, bridging these different medical domains.
While these visualizations are mostly a curiosity (the actual search uses the full high-dimensional embeddings), they help validate that our embedding approach makes intuitive sense. When we see trials clustering naturally by disease area and methodology, it suggests our hypothetical documents are successfully projecting queries into the right regions of the embedding space.
From Searching to Matching
Semantic search gets us part of the way there, but trials have complex eligibility requirements that go way beyond what you can capture in a simple search. Even if we find trials that are conceptually related, we need to try to filter out ones that are clearly not a match. That's why I built an agent system on top of the semantic search. Here's how it works:
1. Parse the User Query
The first challenge is making sure we understand the query properly. When someone enters something like "metastatic melanoma with brain metastases," the agent tries to extract and structure the key medical details. This helps ensure we're starting with high-quality inputs before we even begin searching.
2. Run Semantic Search
The agent uses our embedding system to find potentially relevant trials. This gets us a good starting set, but semantic similarity alone can be misleading - a trial for "BRAF-positive melanoma" will appear very similar to one for "BRAF-negative melanoma" in embedding space, even though they're looking for entirely different patients. This is why we need the next steps.
3. Identify Missing Details
Clinical trials often need specific information that most people wouldn't think to include in a search. The agent looks at the eligibility criteria of the most relevant trials and generates targeted questions: "Has the patient had prior immunotherapy?" or "Do you know their BRAF mutation status?" For each question, it analyzes how different answers would affect which trials remain viable matches.
4. Filter Trials by Eligibility
The system shows users both the most relevant trials and the follow-up questions. As users answer these questions, they can see in real-time which trials would be ruled out by their responses. We also show "near miss" trials and explain why they might not be a match, which helps users understand their options better.
Try It
trialsearcher.com is now live and pulls updated data from ClinicalTrials.gov daily. Go try it for free. There’s a small cost to each search, and the cheap Postgres instance I’m using for vector search means performance can be slow. But I’d love to keep it free, so please give it a try and let me know if it’s helpful.
Is it better or worse than your usual way of finding trials?
Any features you’d like to see added?
Interested in a higher-volume usage or integrating it with your own system? Let me know—we can explore hosting options that won’t slow down the public tool.
Final Thoughts
Vitamin C wasn't isolated until 1928, almost 200 years after Lind's scurvy trial. But scurvy was cured long before anyone understood what caused it. It's tempting to think we've progressed beyond this empirical approach, but sometimes throwing things at the wall remains one of the fastest ways to advance therapeutics.
I've had the privilege of talking to some brilliant folks in trials, semantic search, and precision medicine. Their insights helped shape this approach and kept expectations realistic. While LLMs can help with parts of trial matching (like interpreting eligibility text), the full problem remains far from solved and goes much deeper than trial eligibility matching.
Disclaimer: This tool is a proof of concept and not meant to replace professional medical advice. Please consult healthcare professionals before making decisions about clinical trial participation.